Climate mediates continental scale patterns of stream microbial functional diversity
Background
Understanding the large-scale patterns of microbial functional diversity is essential for anticipating climate change impacts on ecosystems worldwide. However, studies of functional biogeography remain scarce for microorganisms, especially in freshwater ecosystems. Here we study 15,289 functional genes of stream biofilm microbes along three elevational gradients in Norway, Spain and China.
Results
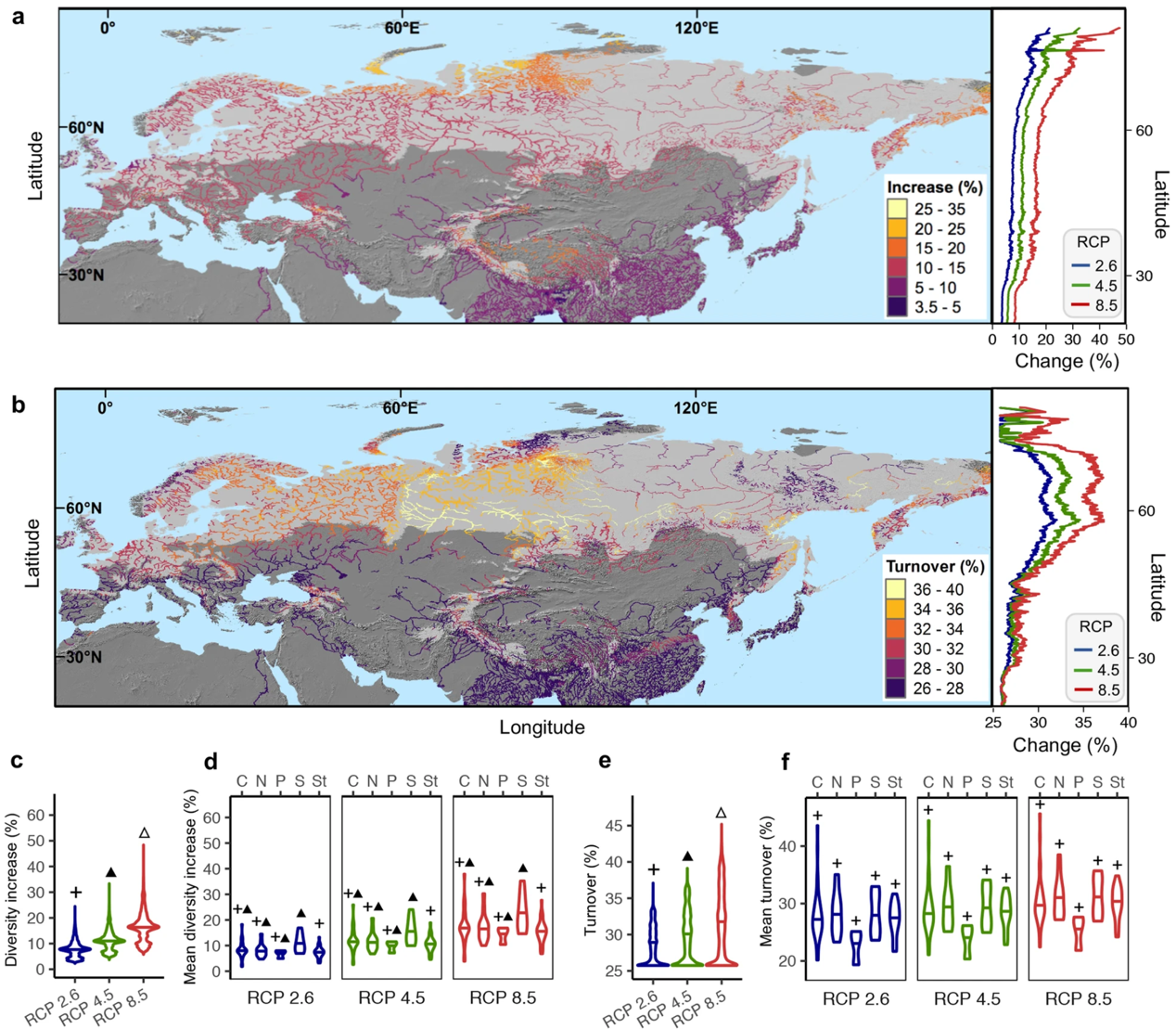
We find that alpha diversity declines towards high elevations and assemblage composition shows increasing turnover with greater elevational distances. These elevational patterns are highly consistent across mountains, kingdoms and functional categories and exhibit the strongest trends in China due to its largest environmental gradients. Across mountains, functional gene assemblages differ in alpha diversity and composition between the mountains in Europe and Asia. Climate, such as mean temperature of the warmest quarter or mean precipitation of the coldest quarter, is the best predictor of alpha diversity and assemblage composition at both mountain and continental scales, with local non-climatic predictors gaining more importance at mountain scale. Under future climate, we project substantial variations in alpha diversity and assemblage composition across the Eurasian river network, primarily occurring in northern and central regions, respectively.
Conclusions
We conclude that climate controls microbial functional gene diversity in streams at large spatial scales; therefore, the underlying ecosystem processes are highly sensitive to climate variations, especially at high latitudes. This biogeographical framework for microbial functional diversity serves as a baseline to anticipate ecosystem responses and biogeochemical feedback to ongoing climate change.
作者:
Félix Picazo, Annika Vilmi, Juha Aalto, Janne Soininen, Emilio O. Casamayor, Yongqin Liu, Qinglong Wu, Lijuan Ren, Jizhong Zhou, Ji Shen & Jianjun Wang*. Microbiome. 2020. DOI:10.1186/s40168-020-00873-2
